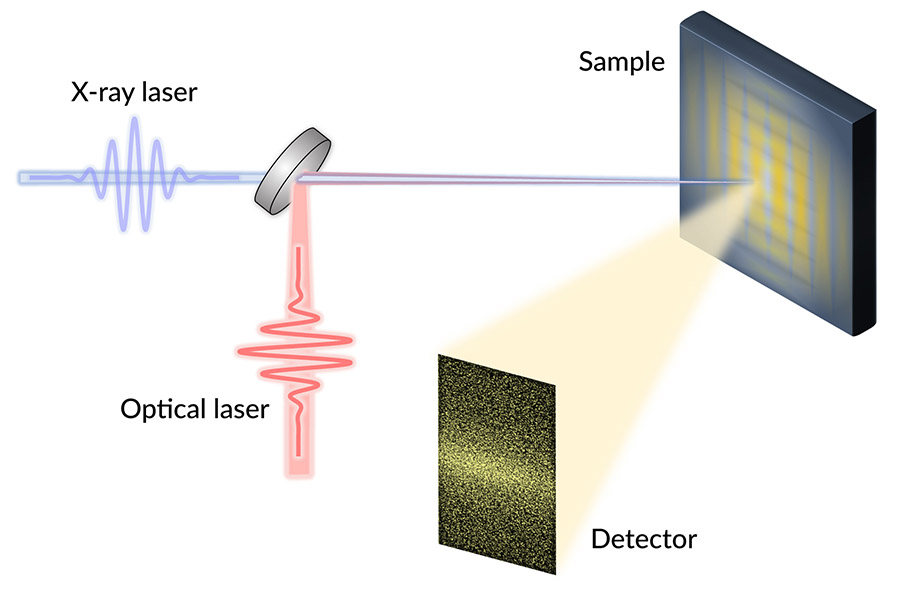
Room-temperature superconductors could transform everything from electrical grids to particle accelerators to computers – but before they can be realized, researchers need to better understand how existing high-temperature superconductors work.
Now, researchers from the Department of Energy’s SLAC National Accelerator Laboratory, the University of British Columbia, Yale University and others have taken a step in that direction by studying the fast dynamics of a material called yttrium barium copper oxide, or YBCO.
The team reports May 20 in Science that YBCO’s superconductivity is intertwined in unexpected ways with another phenomenon known as charge density waves (CDWs), or ripples in the density of electrons in the material. As the researchers expected, CDWs get stronger when they turned off YBCO’s superconductivity. However, they were surprised to find the CDWs also suddenly became more spatially organized, suggesting superconductivity somehow fundamentally shapes the form of the CDWs at the nanoscale.
“A big part of what we don’t know is the relationship between charge density waves and superconductivity,” said Giacomo Coslovich, a staff scientist at the Department of Energy’s SLAC National Accelerator Laboratory, who led the study. “As one of the cleanest high-temperature superconductors that can be grown, YBCO offers us the opportunity to understand this physics in a very direct way, minimizing the effects of disorder.”
He added, “If we can better understand these materials, we can make new superconductors that work at higher temperatures, enabling many more applications and potentially addressing a lot of societal challenges – from climate change to energy efficiency to availability of fresh water.”
Observing fast dynamics
The researchers studied YBCO’s dynamics at SLAC’s Linac Coherent Light Source (LCLS) X-ray laser. They switched off superconductivity in the YBCO samples with infrared laser pulses, and then bounced X-ray pulses off those samples. For each shot of X-rays, the team pieced together a kind of snapshot of the CDWs’ electron ripples. By pasting those together, they recreated the CDWs rapid evolution.
“We did these experiments at the LCLS because we needed ultrashort pulses of X-rays, which can be made at very few places in the world. And we also needed soft X-rays, which have longer wavelengths than typical X-rays, to directly detect the CDWs,” said staff scientist and study co-author Joshua Turner, who is also a researcher at the Stanford Institute for Materials and Energy Sciences. “Plus, the people at LCLS are really great to work with.”
These LCLS runs generated terabytes of data, a challenge for processing. “Using many hours of supercomputing time, LCLS beamline scientists binned our huge amounts of data into a more manageable form so our algorithms could extract the feature characteristics,” said MengXing (Ketty) Na, a University of British Columbia graduate student and co-author on the project.
The team found that charge density waves within the YBCO samples became more correlated – that is, more electron ripples were periodic or spatially synchronized – after lasers switched off the superconductivity.
“Doubling the number of waves that are correlated with just a flash of light is quite remarkable, because light typically would produce the opposite effect. We can use light to completely disorder the charge density waves if we push too hard,” Coslovich said.
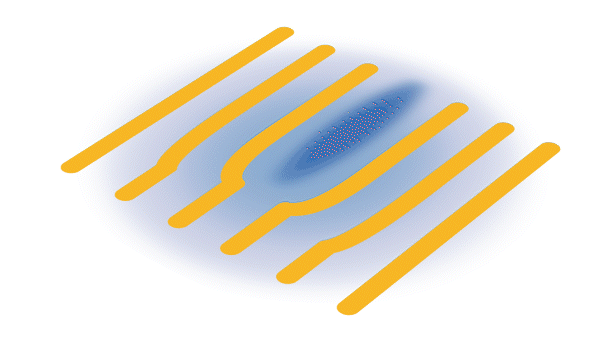
To explain these experimental observations, the researchers then modeled how regions of CDWs and superconductivity ought to interact given a variety of underlying assumptions about how YBCO works. For example, their initial model assumed that a uniform region of superconductivity when shut off with light would become a uniform CDW region – but of course that didn’t agree with their results.
“The model that best fits our data so far indicates that superconductivity is acting like a defect within a pattern of the waves. This suggests that superconductivity and charge density waves like to be arranged in a very specific, nanoscopic way,” explained Coslovich. “They are intertwined orders at the length scale of the waves themselves.”
Illuminating the future
Coslovich said that being able to turn superconductivity off with light pulses was a significant advance, enabling observations on the time scale of less than a trillionth of a second, with major advantages over previous approaches.
“When you use other methods, like applying a high magnetic field, you have to wait a long time before making measurements, so CDWs rearrange around disorder and other phenomena can take place in the sample,” he said. “Using light allowed us to show this is an intrinsic effect, a real connection between superconductivity and charge density waves.”
The research team is excited to expand on this pivotal work, Turner said. First, they want to study how the CDWs become more organized when the superconductivity is shut off with light. They are also planning to tune the laser’s wavelength or polarization in future LCLS experiments in hopes of also using light to enhance, instead of quench, the superconducting state, so they could readily turn the superconducting state off and on.
“There is an overall interest in trying to do this with pulses of light on very fast timescales, because that can potentially lead to the development of superconducting, light-controlled devices for the new generation of electronics and computing,” said Coslovich. “Ultimately, this work can also help guide people who are trying to build room-temperature superconductors.”
This research is part of a collaboration between researchers from LCLS, SLAC’s Stanford Synchrotron Radiation Lightsource (SSRL), UBC, Yale University, the Institut National de la Recherche Scientifique in Canada, North Carolina State University, Universita CAattolica di Brescia and other institutions. This work was funded in part by the DOE Office of Science. LCLS and SSRL are DOE Office of Science user facilities.
Citation: Scott Wandel et al., Science, 20 May 2022 (10.1126/science.abd7213)
This is a reposting of my news feature courtesy of Stanford Linear Accelerator Laboratory.








